Our research fosters a synergy between studies of nonribosomal peptide synthetases and NMR method developments. The high significance of NRPSs provides a strong motivation to overcome experimental challenges and hence develop new methods. NRPSs are microbial enzymatic assembly lines that produce important natural products that confer virulence to pathogens (Vibrio cholera, Mycobacterium tuberculosis, Yersinia pestis-our system) or that are valuable therapeutics, including antibiotics (penicillin, bacitracin), antitumor agents (bleomycin, epothilone), and immunosuppressants (rapamycin). Understanding NRPS synthesis may not only help combat virulence but also produce novel pharmaceuticals. Our immediate goal is to determine the molecular mechanism for the interplay between NRPS chemistry and domain communication, while determining uncharacterized NRPS domain structures. Simultaneously, we develop NMR methods to overcome challenges (complexity of NMR spectra, sensitivity losses) or to provide a more complete description of molecular events. The new methods then provide access to NMR studies of NRPS domains heretofore unamenable for the technique. Our overarching goal is to provide a molecular basis for silencing or reprogramming of NRPSs.
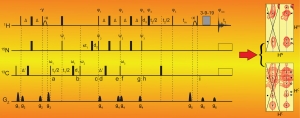
NRPSs use multiple domains, organized in contiguous modules, to covalently load, modify, and join simple substrates in an assembly line fashion and produce complex secondary metabolites. Substrates are tethered to a phosphopantetheine arm post-translationally introduced on a thiolation domain (T) that visits partner catalytic domains during synthesis. This remarkable organization holds the promise of producing novel pharmaceuticals by swapping domains or modules to reprogram NRPS assembly lines. However, many NRPS domain interactions remain uncharacterized, the structure and mechanism of important domains are unknown, and artificially engineered NRPSs are generally unproductive. Moreover, NRPSs are subject to both inter- and intra-domain dynamics. Thus, NRPSs are not rigid assemblies but undergo a series of sequential, transient domain interactions to perform successive catalytic steps. This finding poses new challenges to NRPS engineering or, in general, to understanding the biosynthesis of their natural products. To understand NRPS domain communication, it is not sufficient to identify binding surfaces between domains or determine the structure of multidomain units. One must also characterize the energetics, kinetics, and dynamics involved in competing molecular interactions and unravel their molecular mechanisms. To this aim, we use a combination of NMR studies, biochemistry, and biophysical techniques, and we apply them to various combinations of NRPS domains.
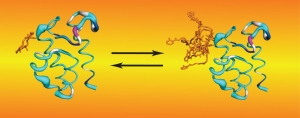
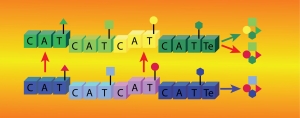
The following example illustrates the unique information NMR studies can provide. We have found that NRPS thiolation domains (T) interact with their phosphopantetheine arms, and we have demonstrated that substrate loading alters this interaction in a manner that likely modulates the affinity between T domains and their binding partners. Rapid hydrolysis of tethered substrates had impeded structural studies of loaded carrier proteins. We overcame this obstacle by performing biochemistry in our NMR tubes, under conditions that convert T domains to their loaded forms and maintain them in that form during NMR experiments. This procedure also permitted for controlled incorporation of NMR isotopes and allowed us to probe for the dynamics of the phosphopantetheine arm before and after substrate loading. We found that, although they interact with T domains in a well-defined manner, as evidenced by our solution structures, prosthetic groups do so in a transient manner. Thus, they transition between states where they extend in solution in a disordered manner and states where they bind to the core carrier protein. Both states may be required at different moments during NRPS synthesis. All these observations have led to new hypotheses that are currently probed in our lab through a variety of experiments and methods.
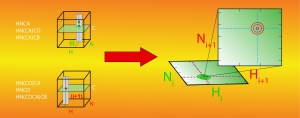
We develop new experimental and computational NMR methods. For example, we have designed a mathematical procedure to facilitate resonance assignments in NMR studies. Traditional approaches consist in identifying spectroscopic patterns within a series of 3D NMR spectra and subsequently combining the information in hope of finding a unique solution. This task is arduous and demanding for large proteins, and we have translated it into mathematical operations applied to NMR spectra. We use covariance to identify patterns in pairs of spectra and element-wise matrix product to combine information from different pairs of spectra. The outcome is a 4D correlation map that combines all information otherwise provided separately by 3D spectra. With this correlation map, for each (H,N) amide correlation in an HN-HSQC, a second HN-HSQC reveals the signals of successor or predecessor amide groups. The procedure greatly facilitates backbone resonance assignment, and a variant was designed for methyl resonance assignments. To facilitate the accurate determination of distance constraints, we developed a pulse sequence enabling the simultaneous acquisition of 15N- and 13C-edited NOESY spectra through so-called non-uniform sampling. High resolution spectra can then be measured within a fortieth of that needed for conventional, sequential acquisition. Similar method developments are constantly ongoing in our laboratory as we carry on our challenging studies of NRPS molecular mechanisms.